The term “microbiome” refers to the ecosystem of microorganisms in the human body. It includes microbes in one’s gut, bacteria on the skin, and any microscopic organism that lives within or on any human body part. Interestingly, microorganisms comprising the human microbiome outnumber human cells in the human body they inhabit. Not all microbes are deleterious to human health.
In fact, some are necessary for healthy life.
Microbiome characterization can help microbiome researchers find the link between certain health conditions and populations of microorganisms. The technique could also allow the treatment of previously untreatable diseases through microbiome modification and diagnosis of diseases based on the presence or absence of certain microbial populations in a patient.
For example, non-alcoholic fatty liver disease (NAFLD) usually shows no symptoms and not diagnosed until it has reached an advanced stage. Non-invasive tests for NAFLD can prove to be inconclusive, and a liver biopsy is often needed to confirm the presence of the disease. Liver biopsies, which involve slicing a piece of one’s liver out, has its risks and is not an ideal means of diagnosis. Fortunately, recent developments in the field of microbiome research provides a less risky method of NAFLD detection.
In the case of NAFLD, the microorganisms’ composition in the patient’s stool, which provides an insight to the patient’s gut microbiota, shows potential for predicting the emergence of the disease. Significant differences were observed between bacteria found in stool samples of patients with NAFLD and healthy individuals. It is now possible to predict which individuals have NAFLD with up to 88% accuracy and which patients have advanced NAFLD with up to 94 % accuracy.
Microbiome-based techniques may also allow diagnosis of medical conditions that don’t manifest physical symptoms. For example, populations of certain gut bacteria strains were revealed to have deviated from the normal range in people suffering from chronic fatigue syndrome. Imbalance in gut microbiota compositions have also been linked to depressive illness. Such findings could assist in finding ideal microbiome composition, or compositions, necessary for maintaining physical and mental health.
[blockquote text=”Furthermore, advancements in gene editing technology can extend the diagnostic capability of microbiome-based techniques through genetic engineering of bacterial species that may be part of the microbiome in focus.” text_color=”” width=”” line_height=”undefined” background_color=”” border_color=”” show_quote_icon=”yes” quote_icon_color=””]
Furthermore, advancements in gene editing technology can extend the diagnostic capability of microbiome-based techniques through genetic engineering of bacterial species that may be part of the microbiome in focus. In one instance, engineered E. coliwas used to infiltrate mice gut microbiome to achieve color-coding of their feces. Feces of mice suffering from colitis glowed green due to the presence of the engineered bacteria. The severity of the inflammation can even be correlated to the intensity of the fecal matter glow. Applying this technology to other diseases could make home-based disease diagnosis become a reality sooner rather than later.
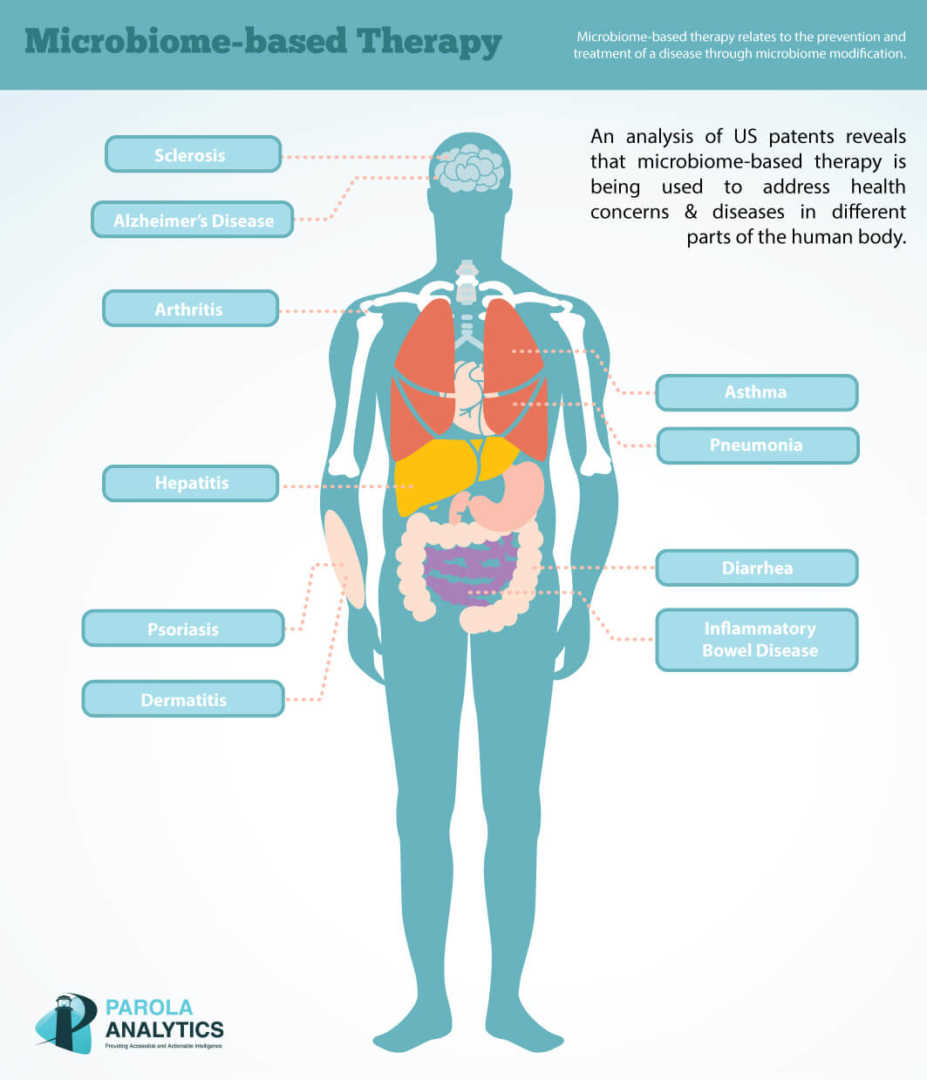
At Parola Analytics, we have analyzed US patents relating to microbiome-based therapy. Our preliminary analysis showed that most patents on microbiome-based therapies are directed to certain diseases, such as hepatitis, diarrhea, psoriasis, and Alzheimer’s disease.
References:
[1] “Nonalcoholic fatty liver disease.” Mayo Clinic. Web. https://www.mayoclinic.org/diseases-conditions/nonalcoholic-fatty-liver-disease/home/ovc-20211638
[2] Ron Sender. “Revised estimates for the number of human and bacteria cells in the body.” PLOS Biology
[3] Ilseung Cho and Martin J. Blaser. “The Human Microbiome: at the interface of health and disease.” Nature review Genetics
[4] Rohit Loomba et. al. “Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease.” Cell Metabolism (2017)
[5] Dorottya Nagy-Szakal et. al. “Fecal metagenomic profiles in subgroups of patients with myalgic encephalomyelitis/chronic fatigue syndrome.” Microbiome (2017)
[6] Sarah Dash, et. al. “The gut microbiome and diet in psychiatry: focus on depression.” Current Opinion in Psychiatry (2015)
[7] Kristina N-M Daeffler et. al. “Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation.” Molecular Systems Biology (2017)